Page Not Found
Page not found. Your pixels are in another canvas.
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas.
About me
This is a page not in th emain menu
Identifying Alternative Splicing effects to Protein interaction Networks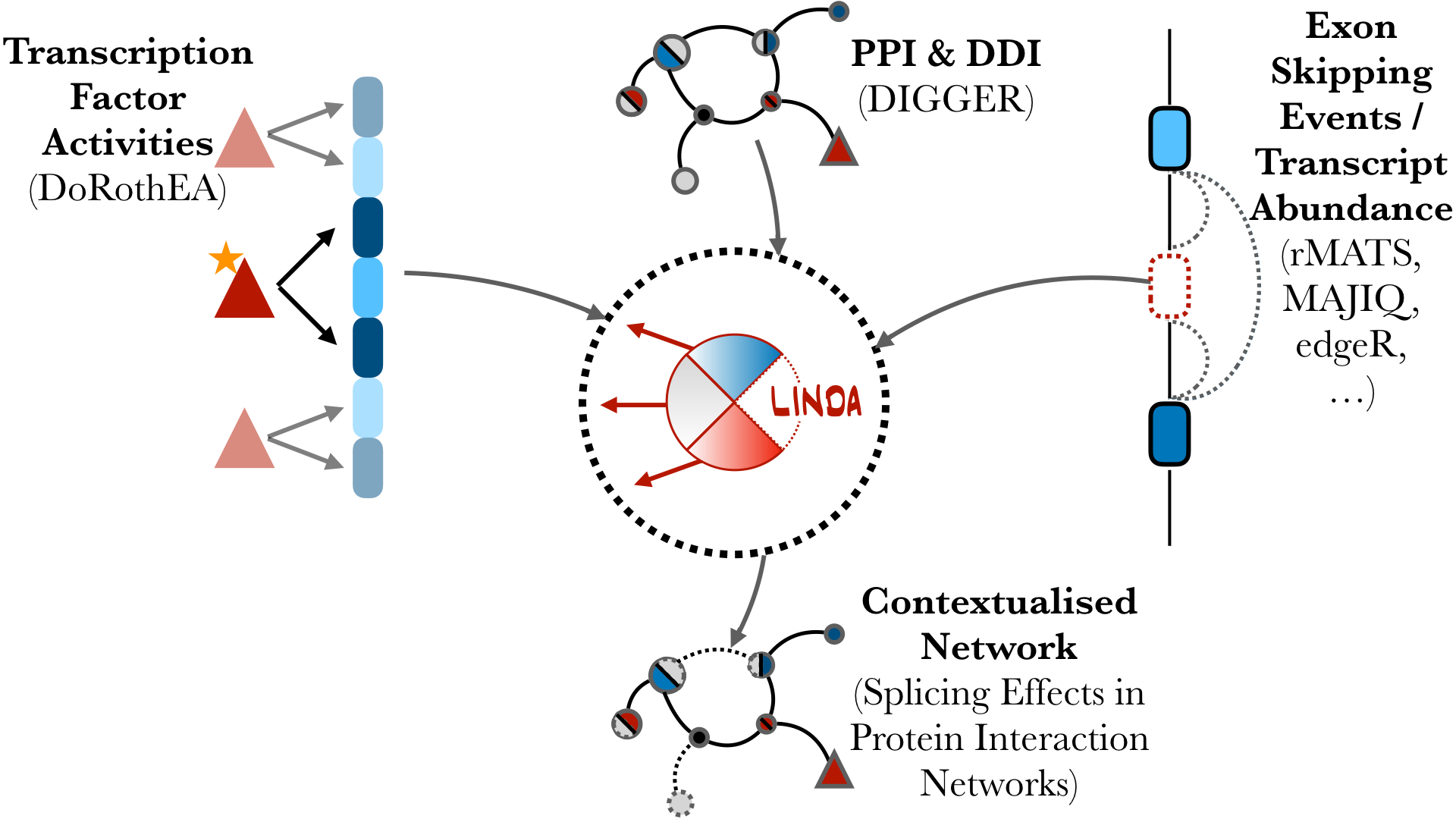
Joint modelling of signalling on multi-cellular systems at a domain resolution data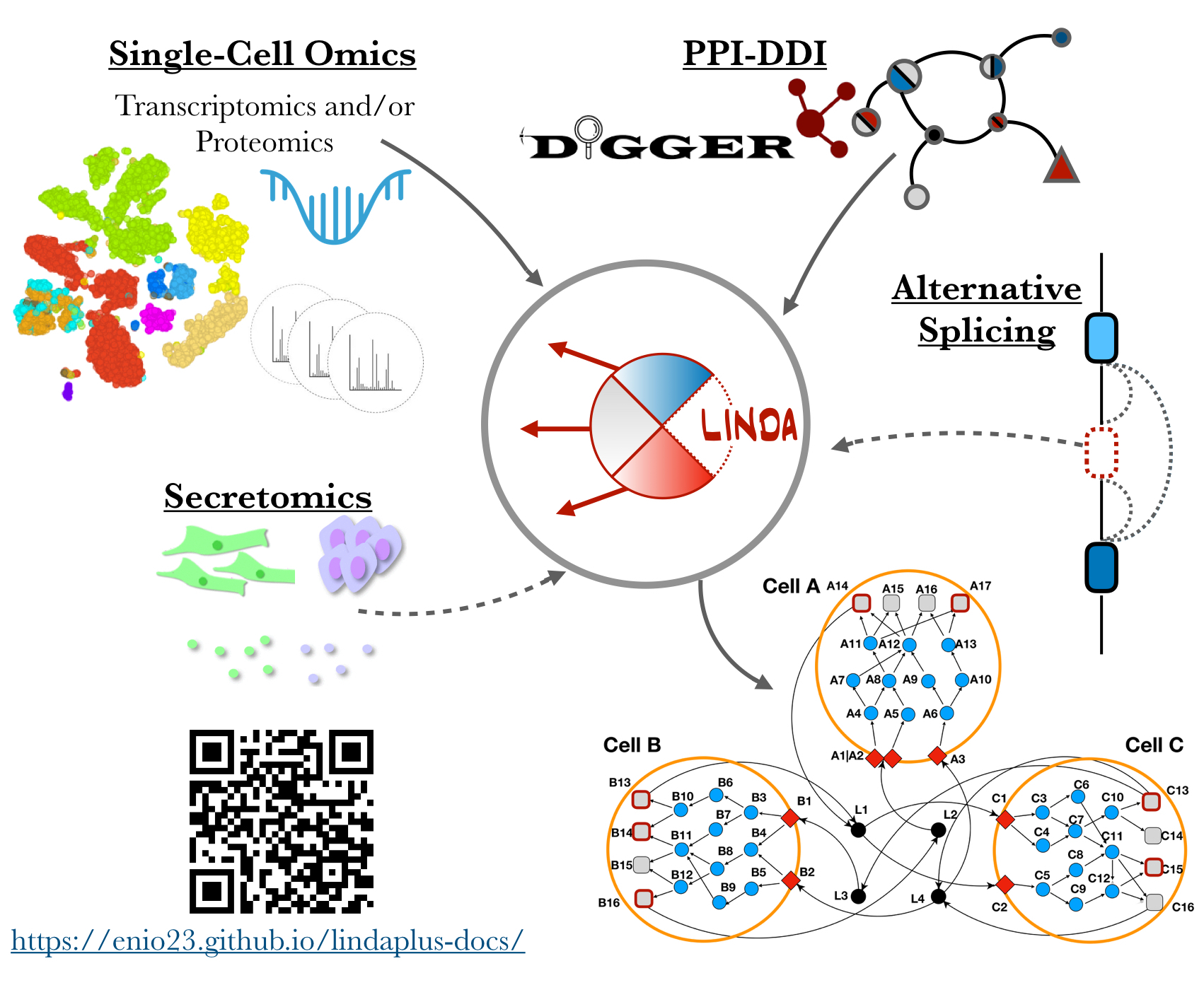
AI and Molecular Dynamics for modelling of signalling on multi-cellular systems at a domain resolution data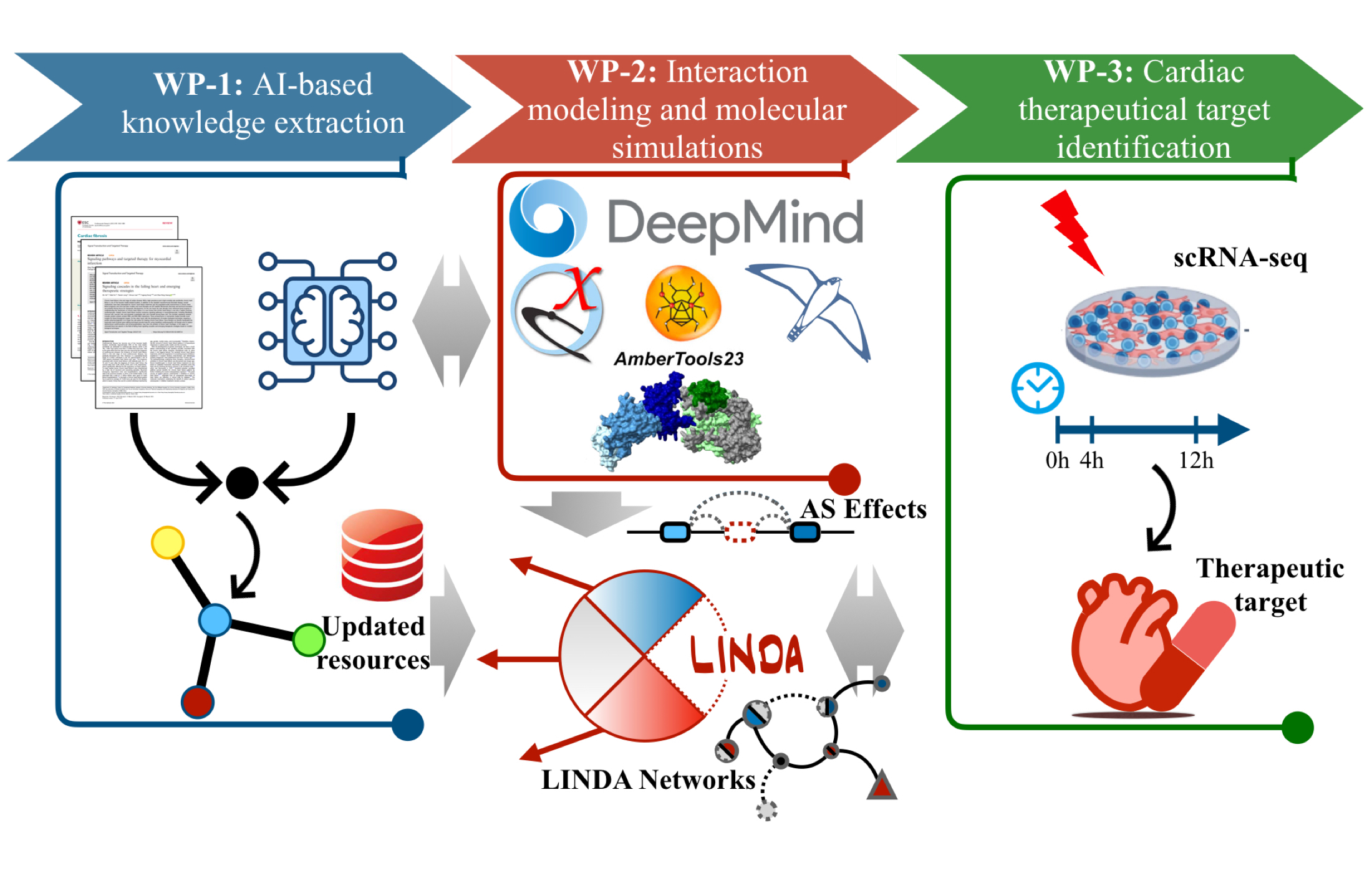
Transverse Aortic COnstriction Multi-omics Analysis (TACOMA) uncovers pathophysiological cardiac molecular mechanisms
Contextualizing large sginalling networks from Gene Expression data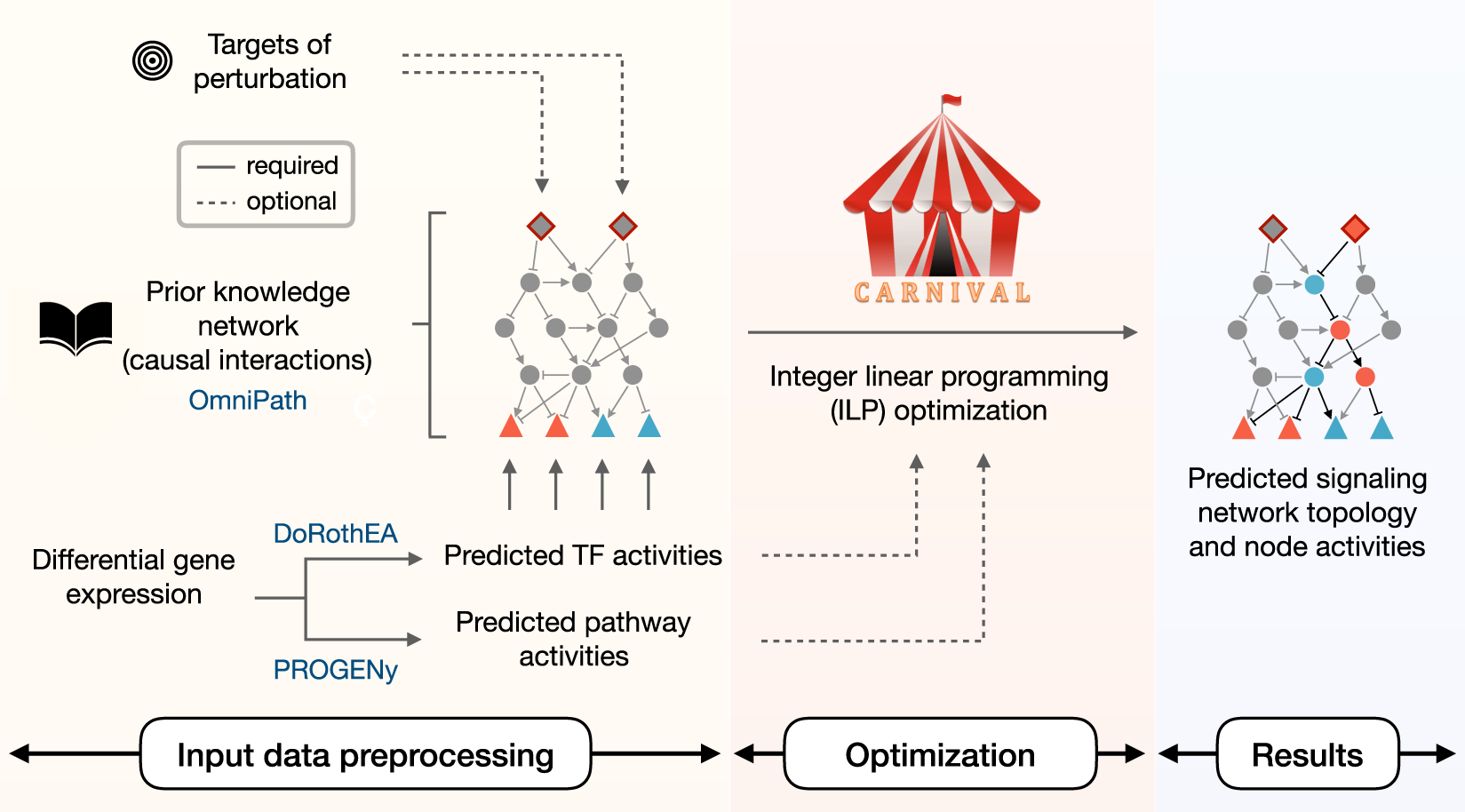
Modelling large-scale signalling networks from shotgun Mass-Spectrometry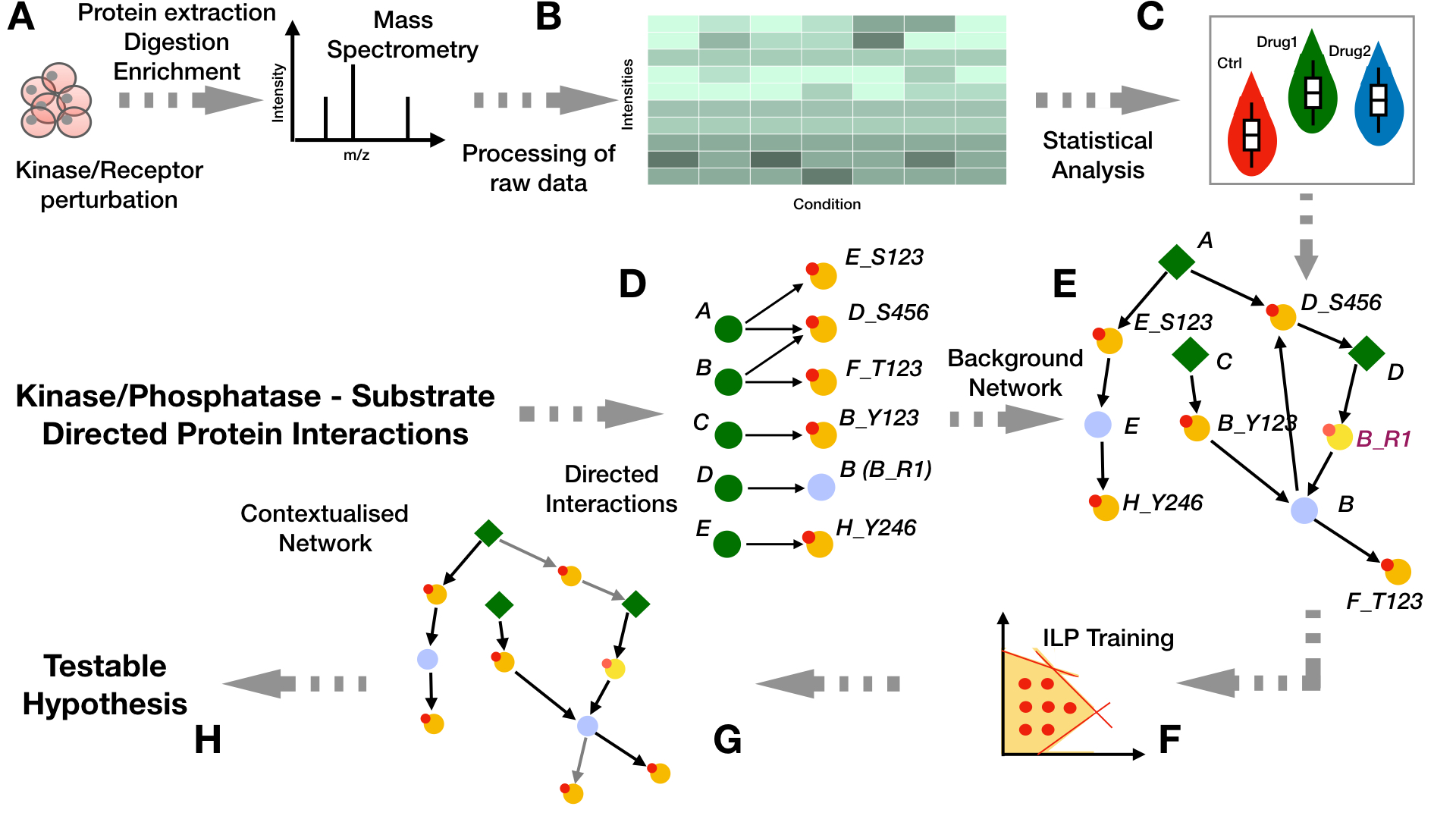
Creating logic-based models of signal transduction networks using different logic formalisms
Published in Molecular Systems Biology, 2019
A comprehensive logic model of endothelin signalling was constructed using phosphoproteomics data from melanoma cell lines, highlighting the role of specific kinases in Endothelin (EDN)-induced cell migration and supporting the strategic selection of kinase inhibitors for combined treatments with EDN receptor antagonists.
Published in IFAC-PapersOnLine, 2019
This paper describes Dynamic-Feeder as a method which combines data-driven inference methods with general literature-based knowledge of proteins interaction networks.
Published in npj Systems Biology and Applications, 2019
CARNIVAL - a causal network contextualization tool which derives network architectures from gene expression footprints.
Published in Bioinformatics, 2020
CellNOpt is a collection of Bioconductor R packages for building logic models from perturbation data and prior knowledge of signalling networks.
Published in Molecular Systems Biology, 2021
COSMOS combines extensive prior knowledge of signaling, metabolic, and gene regulatory networks with computational methods to estimate activities of transcription factors and kinases as well as network‐level causal reasoning.
Published in Journal of Proteome Research, 2021
PHONEMeS is a method that uses high-content shotgun phosphoproteomic data to build logical network models of signal perturbation flow through the implementation of an efficient Integer Linear Programming (ILP) formulation.
Published in Bioinformatics, 2023
LINDA (Linear Integer programming for Network reconstruction using transcriptomics and Differential splicing data Analysis) is a method that integrates resources of protein–protein and domain–domain interactions, transcription factor targets, and differential splicing/transcript analysis to infer splicing-dependent effects on cellular pathways and regulatory networks.
Published in RNA, 2023
Here we show how Adaptive Sampling for direct RNA sequencing with Oxford-Nanopore technology is feasible considering the specific technical and biological challenges.
Published in Database, 2024
Here we present a time-course multi-omics dataset from a mouse model of heart failure induced by transverse aortic constriction (TAC), analyzed using transcriptomics and proteomics to uncover molecular mechanisms of cardiac dysfunction. The data are integrated into TACOMA, a web application enabling interactive exploration of gene and protein expression, alternative splicing, and biological pathway enrichment across disease progression.
Published in BioRxiv, 2025
This study explores the use of open-source large language models (LLMs) for extracting molecular interaction data, specifically protein-protein and bene regulatory relations from scientific literature. The approach shows promising accuracy in relation extraction and offers a toolset for automating biological network curation.
Published:
Conference Proceeding talk at The 31st Annual Intelligent Systems For Molecular Biology and the 22nd Annual European Conference on Computational Biology, where I have presented LINDA.
Published:
Conference talk at the Text Mining: Text Mining for Healthcare and Biology special session of The 33rd Annual Intelligent Systems For Molecular Biology and the 24th Annual European Conference on Computational Biology, where I have presented our work on Evaluating Molecular Relation Extraction using LLMs.
Graduate course, Heidelberg University, 2022
Teaching for the Heidelberg Biosciences International Graduate School (HBIGS). The course focused on the presenting step-by-step analysis of Proteomics data as well as the integrative analysis of Proteomics and Transcriptomics. The lecture was organized over an the EGF-driven protein synthesis case-study date from this paper.
Undergraduate course, Heidelberg University, 2023
Course for the undergraduate medical students of Heidelberg University, teaching basic concepts of programming with R. The course was organized around a simple clinical inflammation data-set provided here.
Graduate course, Heidelberg University, 2023
Course for the Medical Informatics (Profile Bioinformatics). 2022, 2023. In this course I describe the fundamental principles for doing mathematical modelling of Biological System.